Enhanced GAFF Atom Type Assignment
Overview
Accurate atom type assignment is crucial for developing high-quality force fields within the AMBER framework. This precision directly impacts the calculation of bonds, angles, and dihedral values, which ultimately affects the quality of MD simulations. While Antechamber effectively handles atom typing for many common structures, it often encounters difficulties with complex molecular systems, particularly those containing metal atoms or multi-metal frameworks. Incorrect atom typing can lead to inaccurate force field parameters, preventing proper minimization of differences between QM and molecular mechanics (MM) Hessian matrices. easyPARM addresses this limitation with an advanced atom typing module that ensures reliable GAFF atom type assignment even for complex multi-metal-containing structures.
How It Works
Our enhanced atom typing system operates through the following sophisticated process:
1. Molecular Data Analysis
- Parses molecular structure data from MOL2 files, extracting detailed atom properties (element, coordinates) and comprehensive bond information
2. Advanced Structural Analysis
- Ring Identification: Detects and categorizes molecular rings as isolated, fused, bridged, or spiro based on their connectivity patterns
- Hybridization Determination: Calculates the hybridization state (sp, sp², sp³) for each atom using connectivity data and angle information where available
- Aromaticity Detection: Implements a comprehensive algorithm for identifying aromatic systems, including challenging cases involving heteroatoms, metal coordination, and fused ring structures
- Bridge Atom Recognition: Identifies bridge atoms in structures like biphenyl derivatives
- Chemical Environment Mapping: Characterizes each atom’s complete chemical context based on its connections, ring membership, and association with functional groups
3. Intelligent GAFF Type Assignment
- Assigns appropriate GAFF atom types based on the comprehensive structural analysis, following classification principles from the original GAFF publication and official documentation
Technical Implementation
The module employs several advanced techniques:
Graph-Based Ring Detection
- Utilizes a modified Breadth-First Search (BFS) algorithm with collections.deque to identify all simple cycles up to a specified size
- Applies the Smallest Set of Smallest Rings (SSSR) principle to filter out redundant ring structures
Ring System Classification
- Constructs a ring adjacency graph using defaultdict to map connections between rings
- Classifies ring systems with precise definitions:
- Isolated: Rings with no connections to other rings
- Fused: Rings sharing two or more consecutive atoms (e.g., naphthalene)
- Bridged: Rings connected by non-ring atoms (e.g., biphenyl)
- Spiro: Rings sharing a single atom
- Employs set operations for efficient detection of shared atoms and adjacency determination
Hybridization Determination
- Assigns hybridization states based on atom connectivity patterns
- Implements element-specific rules for C, N, O, S, P and other atoms
- Refines assignments for certain elements using angle data when available
Aromaticity Detection
- Employs a comprehensive algorithm that considers:
- Hückel’s rule (4n+2 π-electrons) with adjustments for ring size and composition
- Heteroatom contributions to π-electron counts
- Metal-coordinated rings and complex fused systems
- Iteratively expands aromatic classification across connected ring systems
GAFF Atom Typing
- Implements a rule-based system following the original GAFF publication Wang et al. (2004)
- References official GAFF documentation, including gaff.dat and gaff2.dat files
- Incorporates classification guidelines from authoritative resources like AMBER atom types guide and AMBER’s official documentation
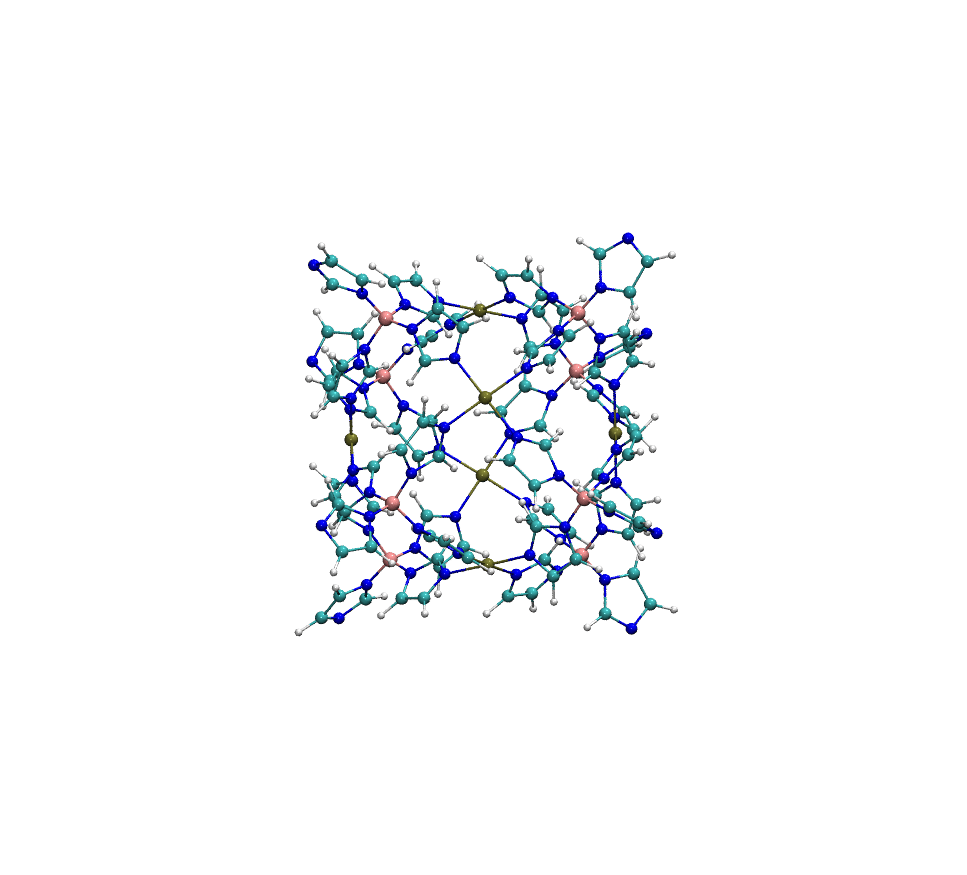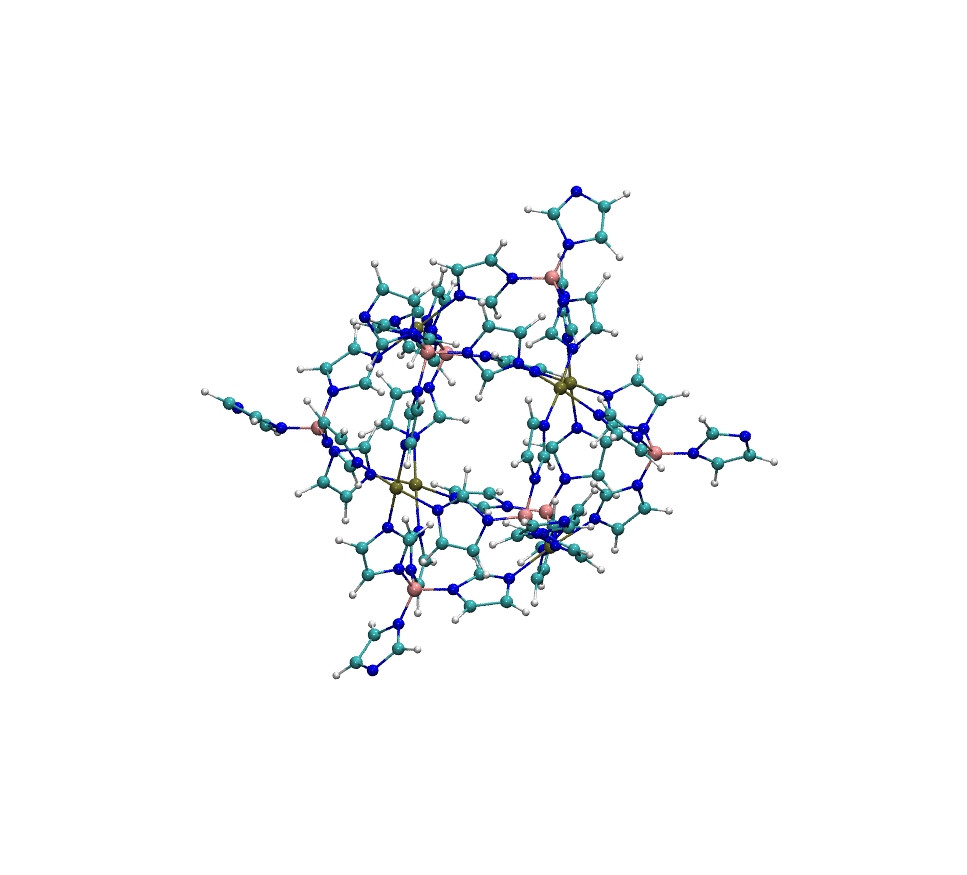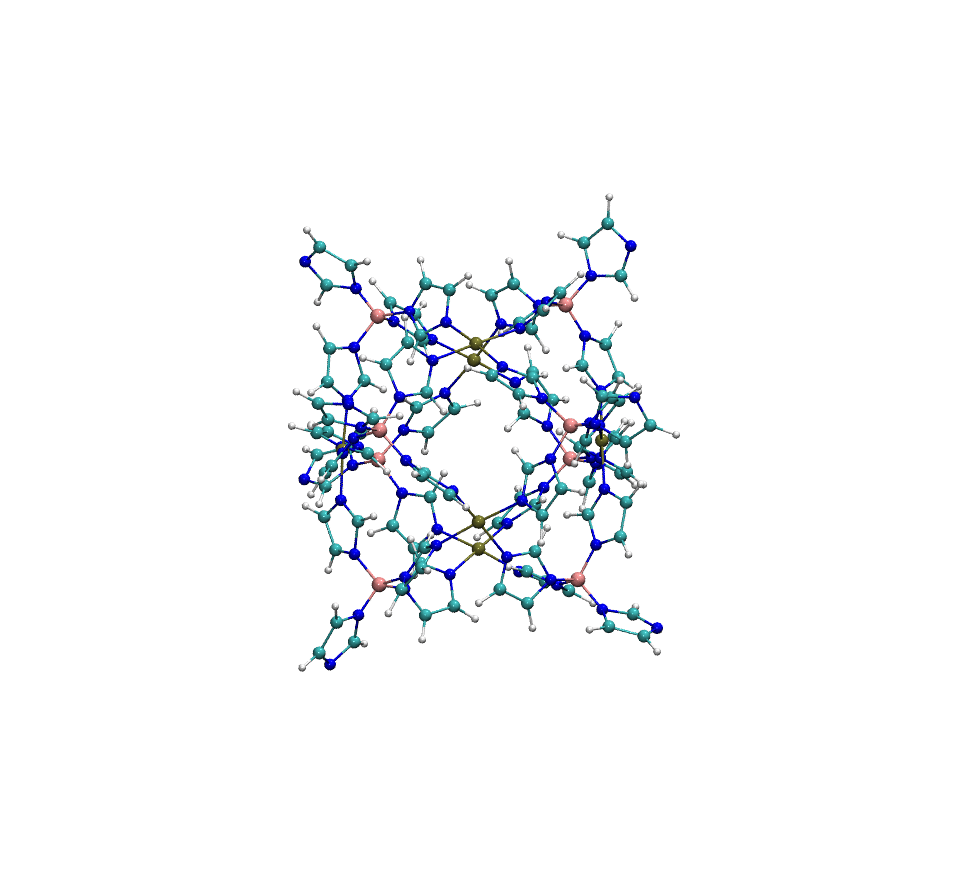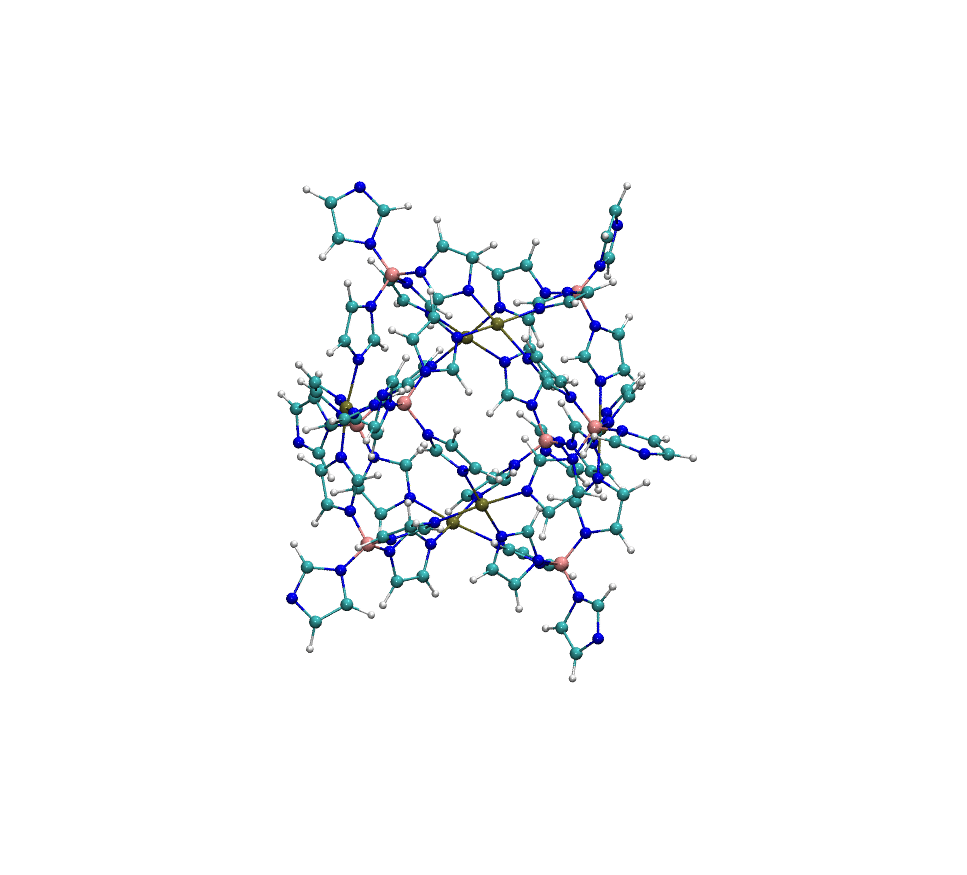
Example: Metal-Organic Polyhedra Case Study
The structure MOP-100 is a representative example of a complex metal-organic polyhedron (MOP), composed of 6 palladium (Pd) atoms coordinated with organic ligands. These Pd centers are arranged to form a discrete, cage-like geometry:
When processed with standard Antechamber, it frequently fails to assign atom types correctly. In successful cases, the resulting COMPLEX_antechamber.mol2 file shows overly simplified atom typing, with most carbon atoms assigned as c2 and most nitrogen atoms as n2—a significant oversimplification that compromises force field accuracy. In contrast, easyPARM’s enhanced atom typing system produces COMPLEX_easyPARM.mol2 with properly diversified atom types (cc, cd, etc. for carbons), capturing the true chemical environment of each atom.
This improved atom typing capability significantly enhances force field accuracy for complex systems. particularly those containing multiple metal centers or unusual coordination environments.
MD Simulation